The aim of the project entitled “Approach to structures of difficult protein targets using new mass spectrometry-based methods“, granted in 2015 by the Polish Ministry of Science (MAESTRO 2014/14/A/NZ1/00306), was structural characterization of biologically important protein assemblies selected from a class of protein systems difficult in structural studies, for which classic methods are not applicable or fail. For this aim new, alternative methods, mainly based on mass spectrometry (MS) were proposed, giving hope to overcome many difficulties accompanying the structural studies of such systems. These methods may help to break the gridlock accompanying the studies of important, sometimes basic biological structures caused by the inadequacy of classic methods of structure analysis, like X-ray, NMR or cryo-EM. Specifically, project was subdivided into four tasks specifying classes of targets of interest: (1) intermediate filament proteins with their interactors, (2) kinetochore and centriole subcomplexes, (3) histone pre-mRNA cleavage complex, (4) Aβ oligomers, involved in Alzheimer’s disease, (5) membrane proteins (RAGE receptor, bacterial toxins). These targets represent selected general cellular processes, often involved in major human pathologies. Their structural analysis, important for design of therapeutically beneficial modifiers was often hampered by lack of appropriate analytical tools.

Results. A brief summary of results described in 20 publications is given below, divided into 5 tasks.
Task 1 Intermediate filament (IF) proteins with their interactors.
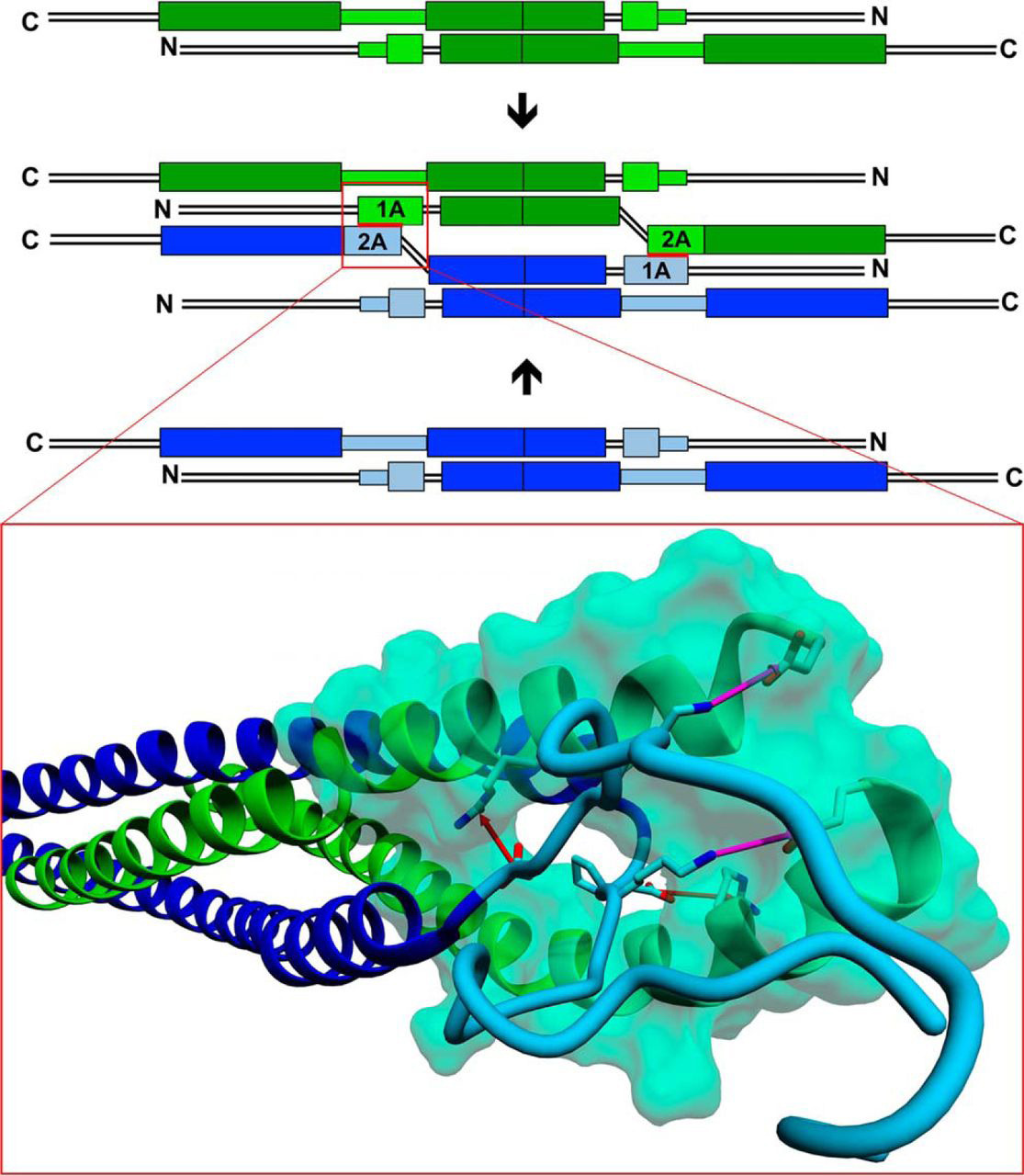
IFs constitute one of the three filamentous systems of the cell skeleton of vertebrates and many invertebrates, accompanying actin/myosin filaments and tubulin microtubules. They not only endow cells with proper mechanical properties but also participate in many cellular processes, also because in vivo filaments assemble and disassemble, move through the cytoplasm in disassembly/assembly cycles and thus participate in the dynamic processes of the cell. The question on IF spatial structure was first asked by W. Astbury in 1930’ties, while contemplating the X-ray patterns of wool/hair keratin (one of the IF proteins) fibers. Therefore the question of IF structure became the first (and thus the oldest) question of molecular biology since this was Astbury who coined the very phrase “molecular biology”. After 90 years knowledge of the IF filament structure is, however, still limited to its subdomain’s structures only, as IF filaments proved to be structurally difficult protein targets. Within the project we have shown, by monitoring the changes in hydrogen-deuterium exchange (HDX) in vimentin, IF protein, tetramers, unit length filaments originating from tetramers by lateral association, and mature filaments, formed by longitudinal ULF assembly, which regions of vimentin are involved in both steps of structural transitions towards filaments. This data allowed us to build a new model for tetramer pairing in ULF and filaments and thus bring closer the final solution of the old but fundamental puzzle.
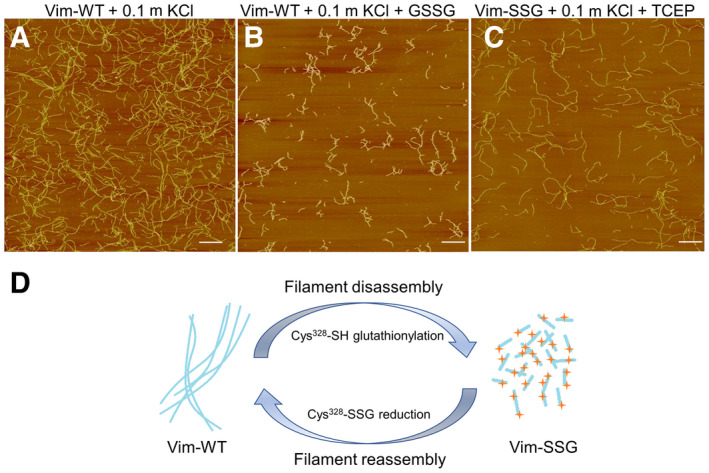
HDX allows to capture the dynamic character of protein structures, so we were also able to monitor the structural changes during filament disassembly/assembly events caused by single point modification of a well-conserved cysteine 328. We have shown that the modification blocks only the elongation step of ULF longitudinal assembly, leaving intact ULF building blocks ready for reassembly after efficient PTM-directed local disassembly and ULF relocation to another localisation within the cell. Vimentin mobility is for instance one of the driving elements of epithelial-to-mesenchymal transition of cancer cells, therefore new knowledge obtained in the project is not only of basic nature.
Some IFs (keratins, vimentin) serve as cancer markers in histopathology. In the analysis of global proteomic experiment of colon adenocarcinoma tissue we have noted that keratin cleavage patterns differ much in cancer and control samples. Further analysis lead us to uncover a new class of cancer-specific peptides, most probably results of neutrophil elastase activity in cancer microenvironment that are possible markers of tumor-infiltrating neutrophil activity, which often correlates with the clinical outcome.
Premchandar A, Mücke N, Poznański J, Wedig T, Kaus-Drobek M, Herrmann H, Dadlez M. Structural Dynamics of the Vimentin Coiled-coil Contact Regions Involved in Filament Assembly as Revealed by Hydrogen-Deuterium Exchange. J Biol Chem. 2016 Nov 25;291(48):24931-24950. doi: 10.1074/jbc.M116.748145. Epub 2016 Sep 30. PMID: 27694444; PMCID: PMC5122765. Kaus-Drobek M, Mücke N, Szczepanowski RH, Wedig T, Czarnocki-Cieciura M, Polakowska M, Herrmann H, Wysłouch-Cieszyńska A, Dadlez M. Vimentin S-glutathionylation at Cys328 inhibits filament elongation and induces severing of mature filaments in vitro. FEBS J. 2020 Dec;287(24):5304-5322. doi: 10.1111/febs.15321. Epub 2020 Apr 21. PMID: 32255262; PMCID: PMC7818121. Kistowski M, Dębski J, Karczmarski J, Paziewska A, Olędzki J, Mikula M, Ostrowski J, Dadlez M. A Strong Neutrophil Elastase Proteolytic Fingerprint Marks the Carcinoma Tumor Proteome. Mol Cell Proteomics. 2017 Feb;16(2):213-227. doi: 10.1074/mcp.M116.058818. Epub 2016 Dec 7. PMID: 27927741; PMCID: PMC5294209.Task 2 Kinetochore and centriole subcomplexes (with D. Glover, Caltech, USA, S. Westermann, Vienna)
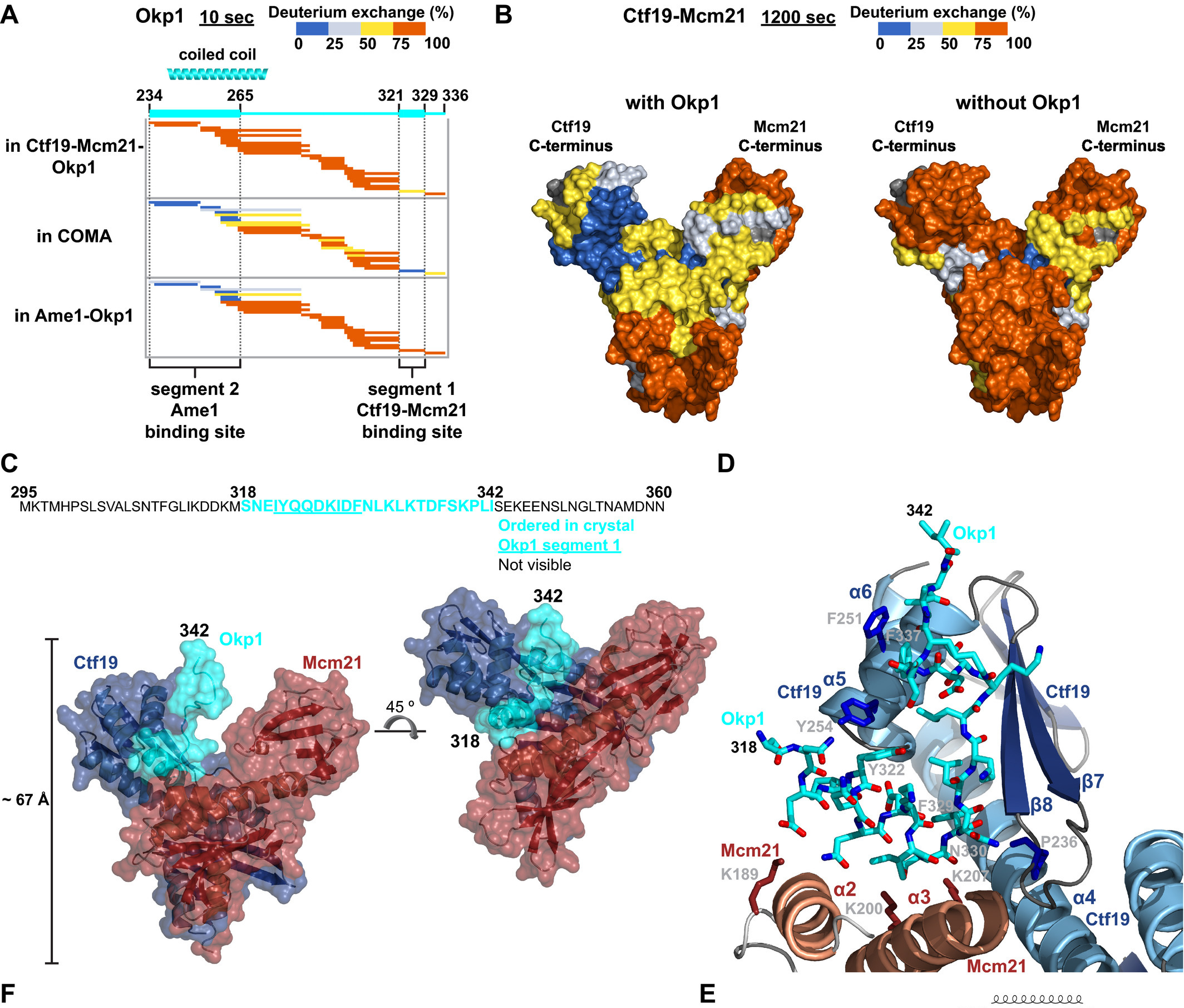
These large protein aggregates orchestrate cell division in most eukaryotic cells. In spite of their fundamental importance their structural characterisation is lagging behind due to their enormous complexity and high content of structurally dynamic regions which makes them intractable by classic methods of structural biology. Alternative methods are thus a must, especially that malfunctions of cell division process are a source of a variety of human diseases. Using HDX we were able to identify regions of contact between several protein components, both of centriolar and kinetochore complexes, therefore uncovering new details on the complex topologies. In case of yeast kinetochore we defined the subunit topology of COMA four-subunits subcomplex in its interaction with inner kinetochore Nkp1-Nkp2 dimer. Figure illustrates the process of identification of contact region of COMA’s Okp1 with Mcm21-Ctf19 dimer (upper left panel) followed by the X-ray analysis of the dimer crystallised in the presence of the identified by HDX Okp1 contact peptide.
For centrioles we first used proteomic approaches to localise a new phosphorylation event in Ana2 that enables further cascade of Plk4-mediated STAN motif phosphorylation, leading to recruitment of Sas6 and initiation of procentriole formation. Following this path we used phosphomimetic form of Ana2 (Ana2-4D) and Sas6 to identify short critical regions required for this interaction, which lie in the C-terminal parts of both proteins. We also confirmed this identification both in vitro and in vivo, for instance by exhaustive mutational studies which allowed to identify critical residues decisive for efficient Sas6 recruitment and procentriole formation.
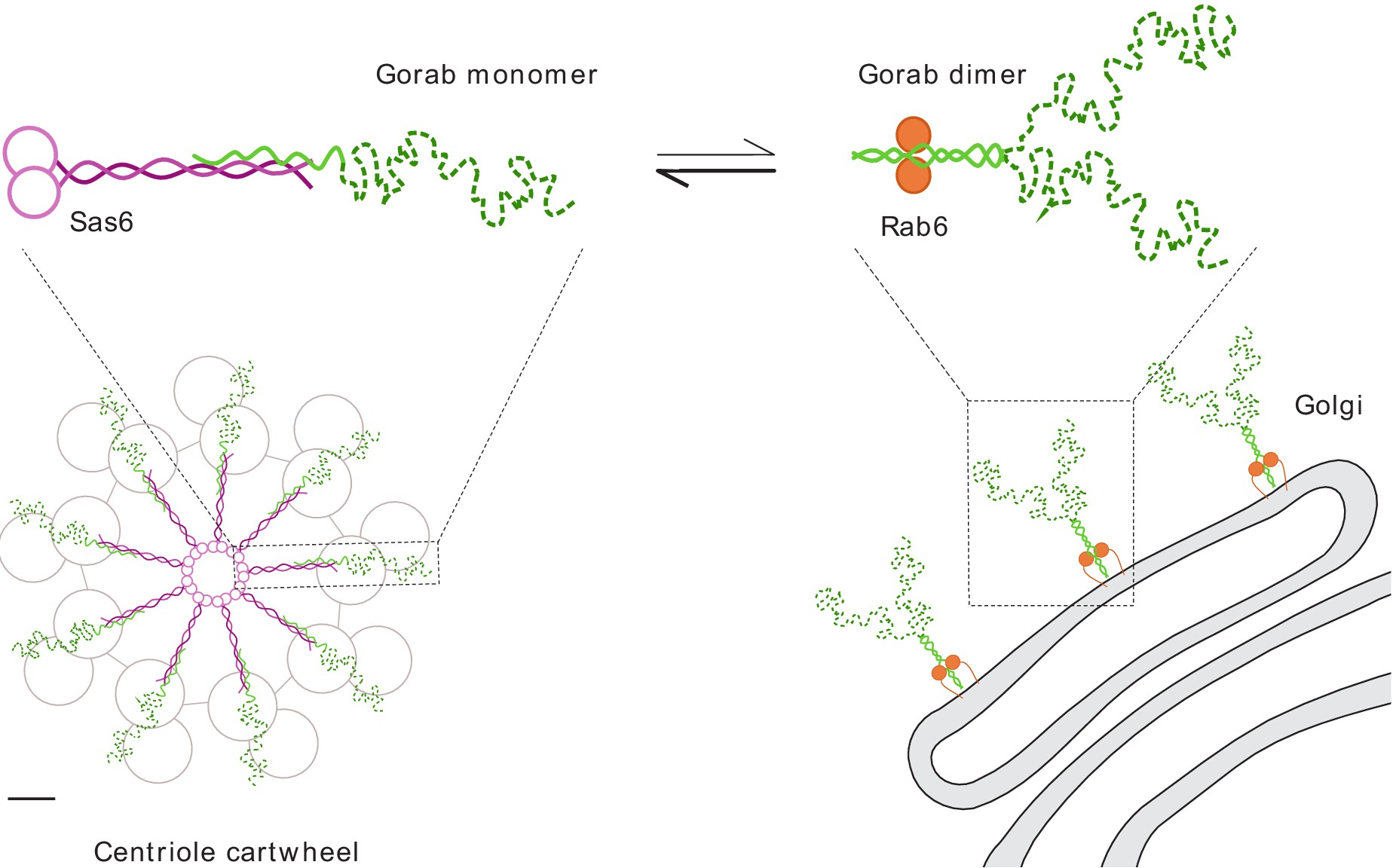
Intrigued by the recent discovery of a new centriole critical protein component – Gorab (protein traditionally known as a localising to trans-Golgi) and equipped with HDX, we aimed at deciphering the interaction network of Gorab, both in centriolar and in Golgi context, and provide structural explanation of its dual role. We identified a core stabilization sequence within Gorab’s C-terminal coiled-coil domain that enables homodimerization, binding to Rab6, and thereby ensures trans-Golgi localization. By contrast, we found that part of the Gorab monomer’s coiled-coil domain undergoes a strong antiparallel interaction with a segment of the parallel coiled-coil dimer of Sas6 which destabilises Gorab dimers and leads to its recruitment to centrioles, instead. Mutation of a single leucine residue in Sas6’s Gorab-binding domain generates a Sas6 variant with a sixteenfold reduced binding affinity for Gorab that cannot support centriole duplication. Thus, Gorab dimers at the Golgi exist in equilibrium with Sas6-associated monomers at the centriole to balance Gorab’s dual role. We have also established that Sas6 binds Ana2 and Gorab in different regions.
Schmitzberger F, Richter MM, Gordiyenko Y, Robinson CV, Dadlez M, Westermann S. Molecular basis for inner kinetochore configuration through RWD domain-peptide interactions. EMBO J. 2017 Dec 1;36(23):3458-3482. doi: 10.15252/embj.201796636. Epub 2017 Oct 18. PMID: 29046335; PMCID: PMC5709738. Fatalska A, Dzhindzhev NS, Dadlez M, Glover DM. Interaction interface in the C-terminal parts of centriole proteins Sas6 and Ana2. Open Biol. 2020 Nov;10(11):200221. doi: 10.1098/rsob.200221. Epub 2020 Nov 11. PMID: 33171067; PMCID: PMC7729032. Fatalska A, Stepinac E, Richter M, Kovacs L, Pietras Z, Puchinger M, Dong G, Dadlez M, Glover DM. The dimeric Golgi protein Gorab binds to Sas6 as a monomer to mediate centriole duplication. Elife. 2021 Mar 11;10:e57241. doi: 10.7554/eLife.57241. PMID: 33704067; PMCID: PMC8009671.Task 3 Histone pre-mRNA cleavage complex (with Z. Domiński, North Carolina Univ., USA)
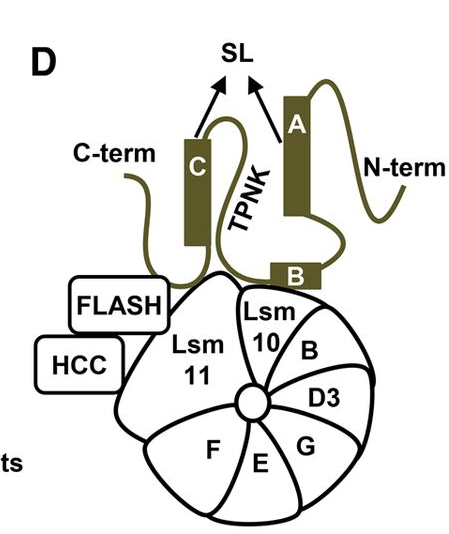
In metazoans, 3′ end processing of replication-dependent histone pre-mRNAs occurs through a specialized 3′ end processing reaction, unique for histone mRNAs. Expression of replication-dependent histone genes is restricted to S-phase of the cell cycle, occurring concomitantly with DNA replication and providing a bulk of histone proteins for de novo synthesized DNA. Outside of S-phase, replication-dependent histone genes are transcriptionally repressed and their mRNA products are virtually undetectable in the cytoplasm. Histone mRNA processing is carried out by a ribonucleoprotein complex through a single endonucleolytic cleavage, generating mature histone mRNAs that lack a poly(A) tail. It requires the multi-subunit U7 snRNP, FLICE-associated huge protein (FLASH), the stem–loop binding protein (SLBP) and histone pre-mRNA cleavage complex (HCC), consisting of several polyadenylation factors. The exact composition of the U7 snRNP, complex topology and details of SLBP and FLASH function in processing remain unclear. In the project we first used proteomic approaches to identify components of Drosophila and mouse U7 snRNP and demonstrated that reconstituted semi-recombinant holo U7 snRNP has the same composition and functional properties as endogenous U7 snRNP, and accurately cleaves histone pre-mRNAs in a reconstituted in vitro processing reaction.
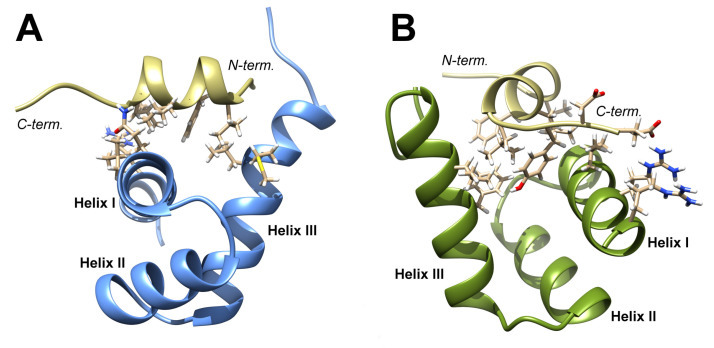
Transcriptional activity of all classes of replication-dependent histone genes is controlled by Nuclear Protein, Ataxia-Telangiectasia Locus (NPAT). NPAT also co-localizes with U7 snRNP and with FLASH through its distant C terminus. We added new details about topology of the histone mRNA processing complex by demonstrating that FLASH plays two roles in 3' end processing of histone pre-mRNAs: it interacts with Lsm11 in the same region that binds NPAT, and it cooperates with SLBP to recruit U7 snRNP to histone pre-mRNA (upper panel). To obtain molecular details of NPAT-FLASH interaction we used multidimensional NMR and in silico modeling to analyze the C-terminal domain in FLASH and YARP in an unbound form and in a complex with the last 31 amino acids of NPAT. Finally we have built a molecular model of the complex of appropriate fragments of both proteins (lower panel).
Skrajna A, Yang XC, Dadlez M, Marzluff WF, Dominski Z. Protein composition of catalytically active U7-dependent processing complexes assembled on histone pre-mRNA containing biotin and a photo-cleavable linker. Nucleic Acids Res. 2018 May 18;46(9):4752-4770. doi: 10.1093/nar/gky133. PMID: 29529248; PMCID: PMC5961079. Skrajna A, Yang XC, Bucholc K, Zhang J, Hall TMT, Dadlez M, Marzluff WF, Dominski Z. U7 snRNP is recruited to histone pre-mRNA in a FLASH-dependent manner by two separate regions of the stem-loop binding protein. RNA. 2017 Jun;23(6):938-951. doi: 10.1261/rna.060806.117. Epub 2017 Mar 13. Bucholc K, Aik WS, Yang XC, Wang K, Zhou ZH, Dadlez M, Marzluff WF, Tong L, Dominski Z. Composition and processing activity of a semi-recombinant holo U7 snRNP. Nucleic Acids Res. 2020 Feb 20;48(3):1508-1530. doi: 10.1093/nar/gkz1148. PMID: 31819999; PMCID: PMC7026596. Bucholc K, Skrajna A, Adamska K, Yang XC, Krajewski K, Poznański J, Dadlez M, Domiński Z, Zhukov I. Structural Analysis of the SANT/Myb Domain of FLASH and YARP Proteins and Their Complex with the C-Terminal Fragment of NPAT by NMR Spectroscopy and Computer Simulations. Int J Mol Sci. 2020 Jul 24;21(15):5268. doi: 10.3390/ijms21155268. PMID: 32722282; PMCID: PMC7432317.Task 4 Aβ peptide oligomers
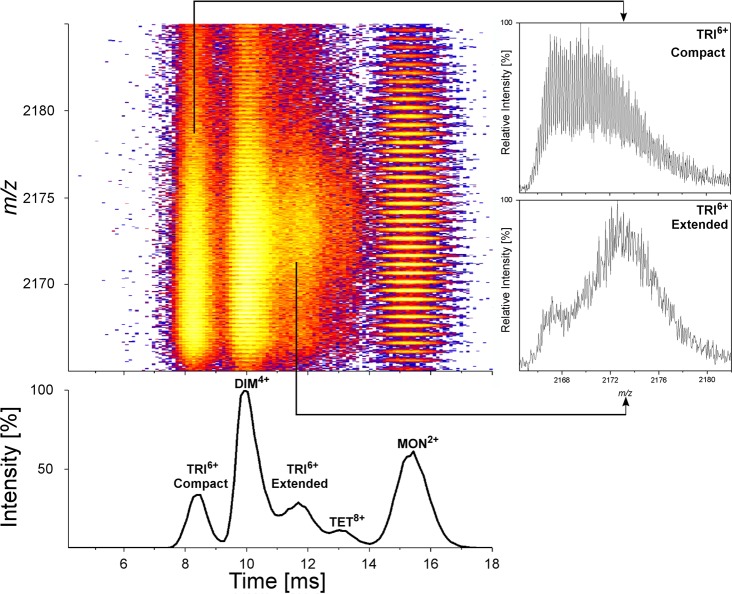
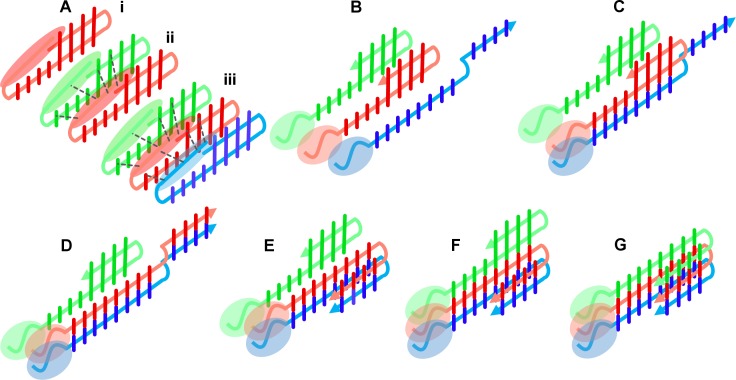
Oligomeric forms of the Aβ 1-40/42 peptide represent the most probable neurotoxic agent in Alzheimer's disease, one of the greatest medical challenges of the present world. The dynamic and heterogeneous character of these oligomers makes their structural characterization by classic methods difficult. Native mass spectrometry, when supported by additional gas phase techniques, like ion mobility separation and hydrogen-deuterium exchange (IM-HDX-MS), enable analysis of different oligomers coexisting in the sample and may provide species-specific structural information for each oligomeric form populated in the gas phase. Fragment of a two dimensional IM-MS spectrum of WT Aβ 1–40 after exchange is shown in the figure to the right. In the project we focused on the role of the N-terminal part of the peptide in shaping the oligomeric equilibria. Previous structural studies have focused on the C-terminal part of Aβ peptide (17-40/42) which provides the core of the amyloid, since nearly all hydrophobic residues with β-structure propensities map to the C-terminal part. The role of the N-terminal region has been neglected, however, it is known for a long time that a peptide variant 17-40/42, deprived of 16 N-terminal residues is a natural product (p3 peptide) of the alternative pathway of amyloid precursor protein cleavage, called “non-amyloidogenic”. Thus, p3, resulting from “non-amyloidogenic” cleavages, contains all “amyloidogenic” properties and all determinants required for fibril assembly, so all neurotoxic properties, believed to result from oligomerization, should thus also be encoded solely within the p3 product. Neurotoxic forms of Aβ peptide are expected to be the result of the amyloidogenic pathway only, and not arising from the non-amyloidogenic pathway. For this reason p3 is believed to be non-neurotoxic, though its role was not studied in much detail. If p3 is non-neurotoxic, while longer 1-40/42 variants are, then the presence of the N-terminus is what makes this peptide pathogenic. Results obtained in the project provided the first experimental evidence for the involvement of the hydrophilic residues from the N-terminal part into a network of interactions stabilizing oligomeric structures. Thus, we obtained structural rationale for the role of N-terminal hydrophilic amino acids of the Aβ peptide (with special focus on three histidine residues H6,H13,H14) in shaping its neurotoxicity and built a molecular model for the transition between an antiparallel stacked-β-hairpin oligomer, suggested previously as an initial oligomeric state, into a parallel β-sheet oligomer, constituting fibrils (Figure to the right).
Przygońska K, Poznański J, Mistarz UH, Rand KD, Dadlez M. Side-chain moieties from the N-terminal region of Aβ are Involved in an oligomer-stabilizing network of interactions. PLoS One. 2018 Aug 6;13(8):e0201761. doi: 10.1371/journal.pone.0201761. PMID: 30080867; PMCID: PMC6078298. Przygońska K, Pacewicz M, Sadowska W, Poznański J, Bal W, Dadlez M. His6, His13, and His14 residues in Aβ 1-40 peptide significantly and specifically affect oligomeric equilibria. Sci Rep. 2019 Jul 1;9(1):9449. doi: 10.1038/s41598-019-45988-1. PMID: 31263161; PMCID: PMC6602940.Task 5 Membrane proteins (RAGE receptor, bacterial toxins)
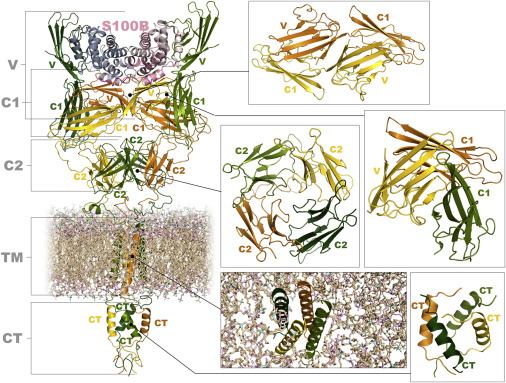
Membrane embedded protein assemblies present a special challenge in structural studies. In the project two such systems were studied, namely RAGE proinflammatory receptor and bacterial toxins. The pattern recognition receptor RAGE (receptor for advanced glycation end-products) transmits proinflammatory signals in several inflammation-related pathological states, including vascular diseases, cancer, neurodegeneration and diabetes. As a crucial mediator of proinflammatory signalling, RAGE activation may enhance a variety of pathophysiological states, including also Alzheimer’s disease. Its oligomerization status is believed to be important in signal transduction, but RAGE oligomeric structures and stoichiometries of non-activated and activated complexes remain unclear. Structural studies revealed only some structures of its truncated, extracellular domains. MS-based techniques used in the present project allowed us to study the structural properties of full length RAGE in lipid-mimicking environment. We have shown the synergy of different domains of RAGE, including its trans-membrane part in shaping oligomeric equilibria, therefore including all parts of proteins into the study seems to be a critical factor for the results to represent the real signal transduction events. Following this we reconstituted the complex of full length RAGE with one of its ligands, S100B, which exhibits neurotoxic activity mediated by the interaction with RAGE, and studied the complex by MS-based methods. Complementing our results with fragmentary structural knowledge on RAGE domains, we constructed a novel molecular model of S100B-RAGE tetramer (Figure to the right).
Moysa A, Hammerschmid D, Szczepanowski RH, Sobott F, Dadlez M. Enhanced oligomerization of full-length RAGE by synergy of the interaction of its domains. Sci Rep. 2019 Dec 30;9(1):20332. doi: 10.1038/s41598-019-56993-9. PMID: 31889156; PMCID: PMC6937306. Moysa A, Steczkiewicz K, Niedzialek D, Hammerschmid D, Zhukova L, Sobott F, Dadlez M. A model of full-length RAGE in complex with S100B. Structure. 2021 Apr 17:S0969-2126(21)00118-0. doi: 10.1016/j.str.2021.04.002. Epub ahead of print. PMID: 33887170.Bacterial toxins: perfringolysin, lysenin.
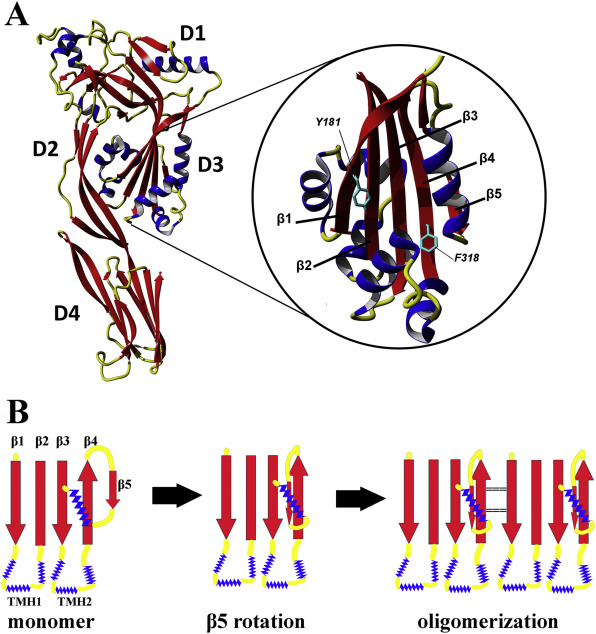
These toxins, produced by Gram-positive bacterial species that are pathogenic in humans and animals, permeabilise membranes of their target cells. Before contact with the membrane toxin molecules are monomeric and soluble, while upon contact they start to oligomerise within the membrane into a multimeric pore structures, destroying the membrane integrity. This is accompanied by a profound structural change and mutual repositioning of its domains. The process is a natural target for therapeutic agents and therefore it is of utmost importance to understand well the molecular details of soluble and membrane bound form and the mechanics of the structural transition. In the project, HDX method provided a detailed insight into structural events of the transition in a series of point mutants used to study the process. We were able to identify, in PFO monomeric state, a structurally strained region of strand 1 decreasing the thermodynamic barrier for β1 and thus enabling the structural conversion during the transition to pore. We also noted a very high sensitivity of the whole toxin structure to single point mutations, leading to profound allosteric changes, spanning distal domains. Therefore point mutations do not yield only local changes but their effects may be global and this should be taken into account while interpreting the results of such studies and constructing the atomic-resolution models of the structural pore transition.
Kulma M, Kacprzyk-Stokowiec A, Traczyk G, Kwiatkowska K, Dadlez M. Fine-tuning of the stability of β-strands by Y181 in perfringolysin O directs the prepore to pore transition. Biochim Biophys Acta Biomembr. 2019 Jan;1861(1):110-122. doi: 10.1016/j.bbamem.2018.08.008. Epub 2018 Aug 21. PMID: 30463694. Kulma M, Kacprzyk-Stokowiec A, Kwiatkowska K, Traczyk G, Sobota A, Dadlez M. R468A mutation in perfringolysin O destabilizes toxin structure and induces membrane fusion. Biochim Biophys Acta Biomembr. 2017 Jun;1859(6):1075-1088. doi: 10.1016/j.bbamem.2017.03.001. Epub 2017 Mar 2. PMID: 28263714.